Often I have a program that I’d like to run with several different settings. For example, I wrote a program in C, compiled into a binary simulate, that computes the amount of gas that a material will adsorb as a function of the pressure of the gas with which the material is in contact. My goal is to make a plot of how much gas a material adsorbs at different pressures, called an adsorption isotherm:
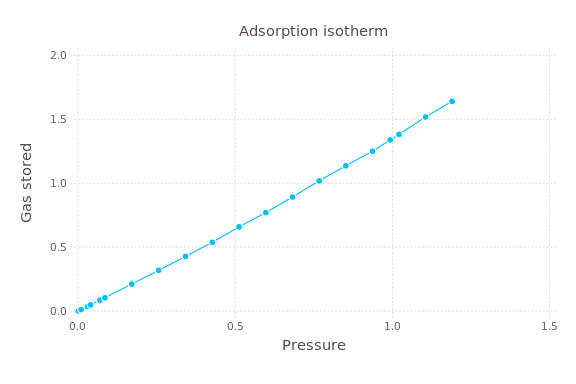
Each point here is an independent simulation at differing gas pressures.
I can compute the quantity of gas a material will adsorb at a particular pressure, say a pressure of 100, by passing the pressure as an argument to the binary of my C program:
./simulate 100A [slow] way to compute an adsorption isotherm is with a Bash script that loops over all pressures:
#!/bin/bash
for P in 1 10 100 1000 10000 100000
do
echo "Computing gas adsorption at pressure $P"
./simulate $P
doneThe problem with this script is that, before I simulate at the pressure at 10, I need to wait for the simulation at the pressure of 1 to be finished. This waiting is not necessary since each simulation is independent of the previous one; the simulation at P=10 does not use any information obtained from the P=1 simulation.
As I have 8 cores on my laptop, I can simulate gas adsorption at these six pressures in parallel by assigning each simulation, differing by the pressure argument, to a different core. This will speed up the rate at which I can compute an adsorption isotherm. The Bash shell has job control to do this. You can start a job in the background by using the ampersand & after a Bash command.
For example, by typing the following into the Bash shell:
./simulate 1 &
./simulate 10 &this will start both P=1 and P=10 simulations on two cores simultaneously. You can see the progress of your jobs by typing jobs into the shell.
Job control is by default only available in the interactive Bash shell, so you must enable job control to use it in a script. Once enabled, this script will start all six simulations at different pressures in the background:
#!/bin/bash
for P in 1 10 100 1000 10000 100000
do
echo "Computing gas adsorption at pressure $P"
./simulate $P &
doneThe ampersand & spawns the job ./simulate $P in the background and allows the loop to continue to the next iteration without waiting for this ./simulate $P job to finish.
This script is dangerous. What if we have 1000 pressures? What if our laptop only has three cores– not enough to assign each of the six pressures to different cores?
The wait command in Bash will wait for all jobs currently running in the background to finish before proceeding in the script. I set up my script to automatically detect the number of cores on my computer, then start one background job for each core, then wait for these jobs to finish before loading the cores with jobs again. This script can handle 1000 pressure points without overloading my computer with simulations.
#!/bin/bash
nprocs=$(cat /proc/cpuinfo | awk '/^processor/{print $3}' | tail -1)
echo "Number of processors: $nprocs"
i=0
for P in 1 10 100 1000 10000 100000
do
echo "Computing gas adsorption at pressure $P"
./simulate $P &
((i++))
[[ $((i%nprocs)) -eq 0 ]] && wait
doneThe last line in the loop tells Bash to wait for all background jobs to finish before continuing to the next iteration in the loop once we have spawned nprocs background jobs.
Can you see an inefficiency in the above script? While much faster than the serial script, this process of submitting jobs suffers from bad load balancing. For nprocs=4, we will submit the jobs for P=1,10,100,1000 and wait for all to finish. But, the simulation at the higher pressure will take much longer than at the lower pressure since there will be more gas molecules in the system. Yet, my program still waits for all four jobs to finish before submitting the next batch; some cores will thus be idle as I am waiting for the P=1000 job to finish.
The solution is not elegant, and I haven’t implemented this yet. This post has a rather complicated Bash code to keep the cores busy by submitting a new job as soon as one of the simulations finish.
UPDATE As pointed out in the comments, GNU Parallel makes running jobs in parallel much easier. You can install GNU Parallel in Ubuntu with sudo apt-get install parallel. Now, running jobs in parallel is as easy as:
#!/bin/bash
nprocs=$(cat /proc/cpuinfo | awk '/^processor/{print $3}' | tail -1)
echo "Number of processors: $nprocs"
# run simulate script with arguments after :::
parallel -j$nprocs ./simulate {} ::: 100000 10000 1000 100 10 1As Daniele Ongari elegantly pointed out in the comments, if possible/known a priori, order the jobs in decreasing run time. For this example, we expect simulations at higher pressures to take the longest since there are more molecules in the system. We want the jobs with the longest run time to begin running immediately. If you don’t clearly see this, consider when we have 10 processors and 19 jobs and, of the 19 jobs, one job takes two hours to run and the remaining jobs each take one hour to run. If we put the two hour job in the beginning, we can keep all processors busy and the entire process will take two hours. On the other hand, if the two hour job is last, the entire process will take three hours; during the last hour, nine of the processors will be idle as we are waiting for the two hour job to finish.
comments powered by Disqus